When it comes sample preparation in bottom-up proteomics
one likes to be as fast, as reproducible and as efficient as possible. Unfortunately,
most of the sample preparations are biased towards certain peptide species. In
this respect hydrophobic proteins, such
as membrane proteins can be troublesome. Also one should consider sample loss
during each preparation step.
However, over the years there have been a
couple of techniques established, that are widely used among proteomics
researchers. Each of them has advantages and disadvantages.
During In-solution protein digestion protein
precipitation in chloroform/methanol is followed by re- solubilization and
digestion in 8M urea. This digestion is achieved in reasonable time compared to
in-gel digestion but has the disadvantage of introducing sample loss during re-solubilization
step.
Second approach is the in-gel preparation,
that follows the idea to entrap the protein solution within a polyacrylamid gel
matrix (usually after SDS PAGE) and subsequently washing out detergents and
performing protein digestion within the
gel. In gel digestion is very time consuming but it is worth though because in
most cases you are ending up with a high number of PSMs.
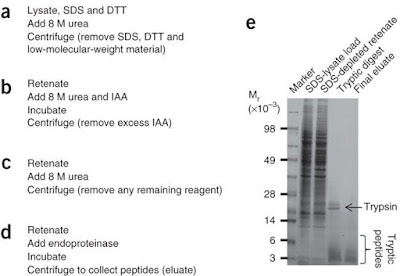
The third technique is called FASP, which
stands for filter aided sample preparation and requires about 7h hands on time.
FASP tries to combined the advantages of the previously mentioned techniques. In
filter aided sample preparation proteins are denatured and kept in solution by
SDS. The SDS-protein mixture is subjected onto a filter cartridge, where all
proteins are bonded. After an SDS-urea exchange, digestion takes place within
the molecular mass cut off filter (be aware of MWCO during selection),
releasing peptides whereas undigested proteins remain within the filter and
would not contaminate the peptide mixture.
Depending on the geometry of the spin filter
and your centrifuge over 50% of the originally used protein amount, ranging
from µg to mg, can be
recovered on peptide level. I found good SDS PAGE from a nature method paper which
served as a control of evaluate the recovery during each step.
A rather new technique is called S-trapping,
the S stands for suspension, because the proteins are trapped within a porous
network made of quartz (SiO2) while being in suspension. Contaminations
and salts have no binding affinity and remain in the flow through. Sample
amounts ranging from ng to µg (read somewhere that 250µg is the maximum protein capacity of the silica
network)
But it all starts with a common SDS step to
solubilize all proteins. Afterwards this SDS micelle is partially broken up and
the proteins begin to become partially denatured. This is when the quartz
networks kicks in and bonds all of these particulate proteins to prevent them
from aggregating with other particulate proteins. Since all the proteins are
attached onto the surface of the network proteolytic digestion enzymes have an
easy job to access all cleavage sites.
Quartz is a good choice since it provides low
metal content (similar to type I silica in HPLC) and low peptide background
during digestion. Additionally, one is able to chemically modify the silica
surface to perform enrichment of certain peptides after digestion (for example
with SDPD, commonly used as crosslinker, for enrichment of cysteine containing
peptides, search C-S-trapping).
A recent study comparing all of these 3 sample preparation approaches indicated that S-trapping outperforms in-solution and FASP in terms
of identification of unique peptides.
The S-Trapping is commercialized by a company
called Protifi. There also provide a unique protease for enhanced b-ion in MSMS
fragementation. Great stuff!